Data analysis with Pandas#
Introduction#
This notebook will give a short introduction to one of the Python data analysis libraries: pandas.
Pandas is widely used in data science, machine learning, scientific computing, and many other data-intensive fields. Some of its advantages are:
data representation: easy to read, suited for data analysis
easy handling of missing data
easy to add/delete columns from
pandasdata structuresdata alignment: intelligent automatic label-based alignment
handling large datasets
powerful grouping of data
native to
Python
Pandas provides rich data structures and indexing functionality to make it easy to reshape, slice and dice, perform aggregations, and select subsets of data. Its key data structures are called the Series and DataFrame.
A Series is a one dimensional array-like object containing an array of data and an associated array of data labels, called its index.
A DataFrame is a two-dimensional tabular, column-oriented data structure with both row and column labels.
import pandas as pd
import numpy as np
Series#
A Series is a 1D array-like object containing an array of data.
first_series = pd.Series([4, 8, -10, np.nan, 2])
first_series
0 4.0
1 8.0
2 -10.0
3 NaN
4 2.0
dtype: float64
The representation of first_series shows the index on the left and the values on the right.
Here the default index format is used: integers 0 to N-1, N being the length of the data.
Creating a Series from a dictionary#
second_series = pd.Series({"Entry1": 0.5, "Entry2": 33.0, "Entry3": 12.0})
second_series
Entry1 0.5
Entry2 33.0
Entry3 12.0
dtype: float64
Creating a Series from a sub-selection of another Series#
third_series = pd.Series(second_series, index=["Entry1", "Entry4"])
third_series
Entry1 0.5
Entry4 NaN
dtype: float64
Adding index names#
first_series.index = ["Row1", "Row2", "Row3", "Row4", "Row5"]
first_series
Row1 4.0
Row2 8.0
Row3 -10.0
Row4 NaN
Row5 2.0
dtype: float64
Accessing information on Series#
first_series
Row1 4.0
Row2 8.0
Row3 -10.0
Row4 NaN
Row5 2.0
dtype: float64
The index object and the values can be accessed individually using:
first_series.index
Index(['Row1', 'Row2', 'Row3', 'Row4', 'Row5'], dtype='object')
first_series.values
array([ 4., 8., -10., nan, 2.])
The index can be used to access the values by a dictionary-like notation or by attribute
first_series.Row1
np.float64(4.0)
first_series["Row1"]
np.float64(4.0)
first_series[["Row3", "Row1"]]
Row3 -10.0
Row1 4.0
dtype: float64
# Change values stored in the Series
first_series["Row4"] = 8
first_series
Row1 4.0
Row2 8.0
Row3 -10.0
Row4 8.0
Row5 2.0
dtype: float64
Filtering, scalar multiplication or mathematical functions can be applied to a Series
# Display all positive values
first_series[first_series > 0]
Row1 4.0
Row2 8.0
Row4 8.0
Row5 2.0
dtype: float64
# Multiply all values in the Series by 2
first_series * 2
Row1 8.0
Row2 16.0
Row3 -20.0
Row4 16.0
Row5 4.0
dtype: float64
# Calculate sine of all values
np.sin(first_series)
Row1 -0.756802
Row2 0.989358
Row3 0.544021
Row4 0.989358
Row5 0.909297
dtype: float64
A Series can also be substituted into many functions that expect a dictionary.
# Check if `Row1` is in the Series
"Row1" in first_series
True
# Check if `Row9` is in the Series
"Row9" in first_series
False
DataFrame#
A DataFrame represents a spreadsheet-like data structure containing an ordered collection of columns.
Each column can be a different value type: numeric, string, boolean, …
Create a DataFrame using a dictionary#
# create dictionary
first_data = {
"Col_1": ["5", 2, "4", "7"],
"Col_2": [
7,
8,
2,
1,
],
"Col_3": [10, 4, 2, 1],
"Col_4": [5, 6, 7, 1],
"Col_5": [9, 9, 2, 1],
"Col_6": [7, 8, 2, 1],
}
# convert dictionary to DataFrame
first_df = pd.DataFrame(first_data)
first_df
| Col_1 | Col_2 | Col_3 | Col_4 | Col_5 | Col_6 | |
|---|---|---|---|---|---|---|
| 0 | 5 | 7 | 10 | 5 | 9 | 7 |
| 1 | 2 | 8 | 4 | 6 | 9 | 8 |
| 2 | 4 | 2 | 2 | 7 | 2 | 2 |
| 3 | 7 | 1 | 1 | 1 | 1 | 1 |
# Display some info about the created DataFrame
first_df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 4 entries, 0 to 3
Data columns (total 6 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Col_1 4 non-null object
1 Col_2 4 non-null int64
2 Col_3 4 non-null int64
3 Col_4 4 non-null int64
4 Col_5 4 non-null int64
5 Col_6 4 non-null int64
dtypes: int64(5), object(1)
memory usage: 324.0+ bytes
first_df.dtypes
Col_1 object
Col_2 int64
Col_3 int64
Col_4 int64
Col_5 int64
Col_6 int64
dtype: object
Note that Col_1 contains a mixture of integers and strings.
Viewing data#
Here is how to display the top and bottom rows of the frame
# display the top 3 rows
first_df.head(3)
| Col_1 | Col_2 | Col_3 | Col_4 | Col_5 | Col_6 | |
|---|---|---|---|---|---|---|
| 0 | 5 | 7 | 10 | 5 | 9 | 7 |
| 1 | 2 | 8 | 4 | 6 | 9 | 8 |
| 2 | 4 | 2 | 2 | 7 | 2 | 2 |
# display the bottom 2 rows
first_df.tail(2)
| Col_1 | Col_2 | Col_3 | Col_4 | Col_5 | Col_6 | |
|---|---|---|---|---|---|---|
| 2 | 4 | 2 | 2 | 7 | 2 | 2 |
| 3 | 7 | 1 | 1 | 1 | 1 | 1 |
Display the index and columns#
first_df.index
RangeIndex(start=0, stop=4, step=1)
first_df.columns
Index(['Col_1', 'Col_2', 'Col_3', 'Col_4', 'Col_5', 'Col_6'], dtype='object')
DataFrame.to_numpy() gives a NumPy representation of the data.
This can be an expensive operation when the DataFrame has columns with different data types.
DataFrame.to_numpy() does not include the index or column labels in the output.
first_df.to_numpy()
array([['5', 7, 10, 5, 9, 7],
[2, 8, 4, 6, 9, 8],
['4', 2, 2, 7, 2, 2],
['7', 1, 1, 1, 1, 1]], dtype=object)
describe() shows a quick statistic summary of the data:
first_df.describe()
| Col_2 | Col_3 | Col_4 | Col_5 | Col_6 | |
|---|---|---|---|---|---|
| count | 4.000000 | 4.000000 | 4.000000 | 4.000000 | 4.000000 |
| mean | 4.500000 | 4.250000 | 4.750000 | 5.250000 | 4.500000 |
| std | 3.511885 | 4.031129 | 2.629956 | 4.349329 | 3.511885 |
| min | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 25% | 1.750000 | 1.750000 | 4.000000 | 1.750000 | 1.750000 |
| 50% | 4.500000 | 3.000000 | 5.500000 | 5.500000 | 4.500000 |
| 75% | 7.250000 | 5.500000 | 6.250000 | 9.000000 | 7.250000 |
| max | 8.000000 | 10.000000 | 7.000000 | 9.000000 | 8.000000 |
# to transpose the data
first_df.T
| 0 | 1 | 2 | 3 | |
|---|---|---|---|---|
| Col_1 | 5 | 2 | 4 | 7 |
| Col_2 | 7 | 8 | 2 | 1 |
| Col_3 | 10 | 4 | 2 | 1 |
| Col_4 | 5 | 6 | 7 | 1 |
| Col_5 | 9 | 9 | 2 | 1 |
| Col_6 | 7 | 8 | 2 | 1 |
Sorting#
# sorting by an axis
first_df.sort_index(axis=1, ascending=False)
| Col_6 | Col_5 | Col_4 | Col_3 | Col_2 | Col_1 | |
|---|---|---|---|---|---|---|
| 0 | 7 | 9 | 5 | 10 | 7 | 5 |
| 1 | 8 | 9 | 6 | 4 | 8 | 2 |
| 2 | 2 | 2 | 7 | 2 | 2 | 4 |
| 3 | 1 | 1 | 1 | 1 | 1 | 7 |
# sorting by values
first_df.sort_values(by=["Col_4"])
| Col_1 | Col_2 | Col_3 | Col_4 | Col_5 | Col_6 | |
|---|---|---|---|---|---|---|
| 3 | 7 | 1 | 1 | 1 | 1 | 1 |
| 0 | 5 | 7 | 10 | 5 | 9 | 7 |
| 1 | 2 | 8 | 4 | 6 | 9 | 8 |
| 2 | 4 | 2 | 2 | 7 | 2 | 2 |
Selection#
Pandas supports several types of multi-axis indexing:
.locto choose rows and columns by label. You have to specify rows and columns based on their row and column labels..ilocto choose rows and columns by position. You have to specify rows and columns by their integer index.
first_df
| Col_1 | Col_2 | Col_3 | Col_4 | Col_5 | Col_6 | |
|---|---|---|---|---|---|---|
| 0 | 5 | 7 | 10 | 5 | 9 | 7 |
| 1 | 2 | 8 | 4 | 6 | 9 | 8 |
| 2 | 4 | 2 | 2 | 7 | 2 | 2 |
| 3 | 7 | 1 | 1 | 1 | 1 | 1 |
Selecting Rows#
# Select index 2 i.e. the 3rd row of the DataFrame
first_df.iloc[2]
Col_1 4
Col_2 2
Col_3 2
Col_4 7
Col_5 2
Col_6 2
Name: 2, dtype: object
If we name the index, this name can also be used to extract data
first_df.index = ["Row1", "Row2", "Row3", "Row4"]
print(f"DataFrame with named index:\n{first_df}")
print(f"\nSelection of the 3rd row:\n {first_df.loc['Row3']}")
DataFrame with named index:
Col_1 Col_2 Col_3 Col_4 Col_5 Col_6
Row1 5 7 10 5 9 7
Row2 2 8 4 6 9 8
Row3 4 2 2 7 2 2
Row4 7 1 1 1 1 1
Selection of the 3rd row:
Col_1 4
Col_2 2
Col_3 2
Col_4 7
Col_5 2
Col_6 2
Name: Row3, dtype: object
# Selecting several rows
# using index
first_df[0:3]
| Col_1 | Col_2 | Col_3 | Col_4 | Col_5 | Col_6 | |
|---|---|---|---|---|---|---|
| Row1 | 5 | 7 | 10 | 5 | 9 | 7 |
| Row2 | 2 | 8 | 4 | 6 | 9 | 8 |
| Row3 | 4 | 2 | 2 | 7 | 2 | 2 |
# using name to select a sequence of rows
first_df.loc["Row1":"Row3"]
| Col_1 | Col_2 | Col_3 | Col_4 | Col_5 | Col_6 | |
|---|---|---|---|---|---|---|
| Row1 | 5 | 7 | 10 | 5 | 9 | 7 |
| Row2 | 2 | 8 | 4 | 6 | 9 | 8 |
| Row3 | 4 | 2 | 2 | 7 | 2 | 2 |
Warning:
Note that contrary to usual Python slices, both the start and the stop are included with loc.
# using name to select rows in a different order
first_df.loc[["Row3", "Row2"]]
| Col_1 | Col_2 | Col_3 | Col_4 | Col_5 | Col_6 | |
|---|---|---|---|---|---|---|
| Row3 | 4 | 2 | 2 | 7 | 2 | 2 |
| Row2 | 2 | 8 | 4 | 6 | 9 | 8 |
# Selecting several rows using their positions
first_df.iloc[:, 2:4]
| Col_3 | Col_4 | |
|---|---|---|
| Row1 | 10 | 5 |
| Row2 | 4 | 6 |
| Row3 | 2 | 7 |
| Row4 | 1 | 1 |
# selecting the 4th row by position
first_df.iloc[3]
Col_1 7
Col_2 1
Col_3 1
Col_4 1
Col_5 1
Col_6 1
Name: Row4, dtype: object
Selecting columns#
# Selecting a single column. The output is a Series.
first_df["Col_4"]
Row1 5
Row2 6
Row3 7
Row4 1
Name: Col_4, dtype: int64
type(first_df["Col_4"])
pandas.core.series.Series
# Equivalent method to select a column
first_df.Col_4
Row1 5
Row2 6
Row3 7
Row4 1
Name: Col_4, dtype: int64
# Selecting several columns using their names
first_df.loc[:, "Col_2":"Col_4"]
| Col_2 | Col_3 | Col_4 | |
|---|---|---|---|
| Row1 | 7 | 10 | 5 |
| Row2 | 8 | 4 | 6 |
| Row3 | 2 | 2 | 7 |
| Row4 | 1 | 1 | 1 |
Selecting subset#
# selecting a subset of the DataFrame using positions
first_df.iloc[1:3, 2:5]
| Col_3 | Col_4 | Col_5 | |
|---|---|---|---|
| Row2 | 4 | 6 | 9 |
| Row3 | 2 | 7 | 2 |
# selecting a subset of the DataFrame using labels
first_df.loc["Row2":"Row3", "Col_3":"Col_5"]
| Col_3 | Col_4 | Col_5 | |
|---|---|---|---|
| Row2 | 4 | 6 | 9 |
| Row3 | 2 | 7 | 2 |
# Getting a single value using labels
first_df.loc["Row3", "Col_4"]
np.int64(7)
# Getting a single value using positions
first_df.iloc[2, 3]
np.int64(7)
# To get faster access to a scalar
first_df.iat[2, 3]
np.int64(7)
Boolean indexing#
# Select the section of DataFrame where the values of `Col_2` are larger than 2
first_df[first_df["Col_2"] > 2]
| Col_1 | Col_2 | Col_3 | Col_4 | Col_5 | Col_6 | |
|---|---|---|---|---|---|---|
| Row1 | 5 | 7 | 10 | 5 | 9 | 7 |
| Row2 | 2 | 8 | 4 | 6 | 9 | 8 |
# Create a new DataFrame
second_df = pd.DataFrame(
np.random.randn(6, 4), index=list("abcdef"), columns=list("ABCD")
)
second_df["E"] = ["one", "one", "three", "two", "three", "four"]
second_df
| A | B | C | D | E | |
|---|---|---|---|---|---|
| a | 0.591110 | 0.222016 | 0.051535 | 1.212998 | one |
| b | 2.178783 | 0.031225 | 1.516338 | -0.133536 | one |
| c | 0.210428 | -1.241545 | 1.606803 | -1.416890 | three |
| d | 1.408970 | -1.014873 | -1.070007 | 0.618123 | two |
| e | 0.061471 | 0.091679 | -0.347482 | -0.030666 | three |
| f | -0.604032 | -0.202534 | -0.946076 | -0.321164 | four |
isin() can also be used for filtering
# Select row if value in 'E' column is 'three'
second_df[second_df["E"].isin(["three"])]
| A | B | C | D | E | |
|---|---|---|---|---|---|
| c | 0.210428 | -1.241545 | 1.606803 | -1.416890 | three |
| e | 0.061471 | 0.091679 | -0.347482 | -0.030666 | three |
Setting#
Adding column to a DataFrame#
Create a Series to be added to second_df.
series_to_add = pd.Series([1, 2, 3, 4, 5, 6], index=["a", "b", "d", "e", "g", "h"])
series_to_add
a 1
b 2
d 3
e 4
g 5
h 6
dtype: int64
second_df["F"] = series_to_add
second_df
| A | B | C | D | E | F | |
|---|---|---|---|---|---|---|
| a | 0.591110 | 0.222016 | 0.051535 | 1.212998 | one | 1.0 |
| b | 2.178783 | 0.031225 | 1.516338 | -0.133536 | one | 2.0 |
| c | 0.210428 | -1.241545 | 1.606803 | -1.416890 | three | NaN |
| d | 1.408970 | -1.014873 | -1.070007 | 0.618123 | two | 3.0 |
| e | 0.061471 | 0.091679 | -0.347482 | -0.030666 | three | 4.0 |
| f | -0.604032 | -0.202534 | -0.946076 | -0.321164 | four | NaN |
Note that the index is aligned: the added series has additional entries g and h and no entries for c and f. The additional entries are discarded in the DataFrame and the absence of entry is marked as NaN.
Setting values by label#
second_df.at["b", "A"] = 0
second_df
| A | B | C | D | E | F | |
|---|---|---|---|---|---|---|
| a | 0.591110 | 0.222016 | 0.051535 | 1.212998 | one | 1.0 |
| b | 0.000000 | 0.031225 | 1.516338 | -0.133536 | one | 2.0 |
| c | 0.210428 | -1.241545 | 1.606803 | -1.416890 | three | NaN |
| d | 1.408970 | -1.014873 | -1.070007 | 0.618123 | two | 3.0 |
| e | 0.061471 | 0.091679 | -0.347482 | -0.030666 | three | 4.0 |
| f | -0.604032 | -0.202534 | -0.946076 | -0.321164 | four | NaN |
Setting values by label#
second_df.iat[1, 0] = 0
second_df
| A | B | C | D | E | F | |
|---|---|---|---|---|---|---|
| a | 0.591110 | 0.222016 | 0.051535 | 1.212998 | one | 1.0 |
| b | 0.000000 | 0.031225 | 1.516338 | -0.133536 | one | 2.0 |
| c | 0.210428 | -1.241545 | 1.606803 | -1.416890 | three | NaN |
| d | 1.408970 | -1.014873 | -1.070007 | 0.618123 | two | 3.0 |
| e | 0.061471 | 0.091679 | -0.347482 | -0.030666 | three | 4.0 |
| f | -0.604032 | -0.202534 | -0.946076 | -0.321164 | four | NaN |
Using a NumPy array#
# Change all values in column 'C'
second_df.loc[:, "C"] = np.arange(len(second_df))
second_df
| A | B | C | D | E | F | |
|---|---|---|---|---|---|---|
| a | 0.591110 | 0.222016 | 0.0 | 1.212998 | one | 1.0 |
| b | 0.000000 | 0.031225 | 1.0 | -0.133536 | one | 2.0 |
| c | 0.210428 | -1.241545 | 2.0 | -1.416890 | three | NaN |
| d | 1.408970 | -1.014873 | 3.0 | 0.618123 | two | 3.0 |
| e | 0.061471 | 0.091679 | 4.0 | -0.030666 | three | 4.0 |
| f | -0.604032 | -0.202534 | 5.0 | -0.321164 | four | NaN |
Missing data#
Pandas uses numpy.nan to represent missing data. This value is by default not included in computations.
Dropping rows with missing data#
The following command will remove rows c and f because they contain one NaN value.
To filter rows containing only NaN values, replace how='any' by how='all'.
second_df.dropna(axis=0, how="any", inplace=False)
| A | B | C | D | E | F | |
|---|---|---|---|---|---|---|
| a | 0.591110 | 0.222016 | 0.0 | 1.212998 | one | 1.0 |
| b | 0.000000 | 0.031225 | 1.0 | -0.133536 | one | 2.0 |
| d | 1.408970 | -1.014873 | 3.0 | 0.618123 | two | 3.0 |
| e | 0.061471 | 0.091679 | 4.0 | -0.030666 | three | 4.0 |
Filling missing data#
The following command replaces NaN by 8.33.
second_df.fillna(value=8.33)
| A | B | C | D | E | F | |
|---|---|---|---|---|---|---|
| a | 0.591110 | 0.222016 | 0.0 | 1.212998 | one | 1.00 |
| b | 0.000000 | 0.031225 | 1.0 | -0.133536 | one | 2.00 |
| c | 0.210428 | -1.241545 | 2.0 | -1.416890 | three | 8.33 |
| d | 1.408970 | -1.014873 | 3.0 | 0.618123 | two | 3.00 |
| e | 0.061471 | 0.091679 | 4.0 | -0.030666 | three | 4.00 |
| f | -0.604032 | -0.202534 | 5.0 | -0.321164 | four | 8.33 |
Define a mask#
True marks the NaN values in the DataFrame.
pd.isna(second_df)
| A | B | C | D | E | F | |
|---|---|---|---|---|---|---|
| a | False | False | False | False | False | False |
| b | False | False | False | False | False | False |
| c | False | False | False | False | False | True |
| d | False | False | False | False | False | False |
| e | False | False | False | False | False | False |
| f | False | False | False | False | False | True |
Some of the methods to deal with missing data#
DataFrame.isna indicates missing values.
DataFrame.notna indicates existing (non-missing) values.
DataFrame.fillna replaces missing values.
Series.dropna drops missing values.
Index.dropna drops missing indices.
Operations#
Statistics#
second_df
| A | B | C | D | E | F | |
|---|---|---|---|---|---|---|
| a | 0.591110 | 0.222016 | 0.0 | 1.212998 | one | 1.0 |
| b | 0.000000 | 0.031225 | 1.0 | -0.133536 | one | 2.0 |
| c | 0.210428 | -1.241545 | 2.0 | -1.416890 | three | NaN |
| d | 1.408970 | -1.014873 | 3.0 | 0.618123 | two | 3.0 |
| e | 0.061471 | 0.091679 | 4.0 | -0.030666 | three | 4.0 |
| f | -0.604032 | -0.202534 | 5.0 | -0.321164 | four | NaN |
# Note that we need to discard column `E`, which contains strings, to calculate the mean
del second_df["E"]
second_df.mean()
A 0.277991
B -0.352339
C 2.500000
D -0.011856
F 2.500000
dtype: float64
Note that NaN values in column F have been automatically discarded.
You can also specify which axis to calculate the mean. For example, to calculate the average for each row,
second_df.mean(1)
a 0.605225
b 0.579538
c -0.112002
d 1.402444
e 1.624497
f 0.968068
dtype: float64
Apply#
Use apply to apply a function along an axis of the DataFrame
third_df = pd.DataFrame({"a": [1, 2, 3, 4, 5], "b": [7, 8, 9, 10, 11]})
third_df
| a | b | |
|---|---|---|
| 0 | 1 | 7 |
| 1 | 2 | 8 |
| 2 | 3 | 9 |
| 3 | 4 | 10 |
| 4 | 5 | 11 |
# calculate the square root of all elements in the DataFrame
third_df.apply(np.sqrt)
| a | b | |
|---|---|---|
| 0 | 1.000000 | 2.645751 |
| 1 | 1.414214 | 2.828427 |
| 2 | 1.732051 | 3.000000 |
| 3 | 2.000000 | 3.162278 |
| 4 | 2.236068 | 3.316625 |
# calculate the sum along one of the axes
third_df.apply(np.sum, axis=0)
a 15
b 45
dtype: int64
third_df.apply(np.sum, axis=1)
0 8
1 10
2 12
3 14
4 16
dtype: int64
Merge#
Pandas provides several tools to easily combine Series and DataFrames.
concatenating objects with concat()#
Syntax:
pd.concat(objs, axis=0, join='outer', ignore_index=False, keys=None, levels=None,
names=None, verify_integrity=False, copy=True)
Simple example:
df1 = pd.DataFrame({"A": [1, 2, 3], "B": [4, 5, 6]})
df2 = pd.DataFrame({"C": [-1, -2, -3], "D": [-4, -5, -6]})
result = pd.concat([df1, df2], join="outer")
result
| A | B | C | D | |
|---|---|---|---|---|
| 0 | 1.0 | 4.0 | NaN | NaN |
| 1 | 2.0 | 5.0 | NaN | NaN |
| 2 | 3.0 | 6.0 | NaN | NaN |
| 0 | NaN | NaN | -1.0 | -4.0 |
| 1 | NaN | NaN | -2.0 | -5.0 |
| 2 | NaN | NaN | -3.0 | -6.0 |
If join='inner', instead of getting the union of the DataFrames,
we will get the intersection. For the 2 example DataFrames, the intersection is empty as shown below:
result = pd.concat([df1, df2], join="inner")
result
| 0 |
|---|
| 1 |
| 2 |
| 0 |
| 1 |
| 2 |
joining with merge()#
Syntax:
pd.merge(left, right, how='inner', on=None, left_on=None, right_on=None,
left_index=False, right_index=False, sort=True,
suffixes=('_x', '_y'), copy=True, indicator=False,
validate=None)
Simple example:
left_df = pd.DataFrame({"key": ["0", "1"], "lval": [1, 2]})
right_df = pd.DataFrame({"key": ["0", "1", "2"], "rval": [3, 4, 5]})
left_df
| key | lval | |
|---|---|---|
| 0 | 0 | 1 |
| 1 | 1 | 2 |
right_df
| key | rval | |
|---|---|---|
| 0 | 0 | 3 |
| 1 | 1 | 4 |
| 2 | 2 | 5 |
pd.merge(left_df, right_df, on="key")
| key | lval | rval | |
|---|---|---|---|
| 0 | 0 | 1 | 3 |
| 1 | 1 | 2 | 4 |
With how='right', only the keys of the right frame are used:
pd.merge(left_df, right_df, on="key", how="right")
| key | lval | rval | |
|---|---|---|---|
| 0 | 0 | 1.0 | 3 |
| 1 | 1 | 2.0 | 4 |
| 2 | 2 | NaN | 5 |
joining on index#
left_df = pd.DataFrame(
{"A": ["A0", "A1", "A2"], "B": ["B0", "B1", "B2"]}, index=["Key0", "Key1", "Key2"]
)
right_df = pd.DataFrame({"C": ["C0", "C2"], "D": ["D0", "D2"]}, index=["Key0", "Key2"])
result = left_df.join(right_df)
result
| A | B | C | D | |
|---|---|---|---|---|
| Key0 | A0 | B0 | C0 | D0 |
| Key1 | A1 | B1 | NaN | NaN |
| Key2 | A2 | B2 | C2 | D2 |
Grouping#
“group by” is referring to a process involving one or more of the following steps:
Splitting data into groups based on some criteria
Applying a function to each group independently
Combining the results into a data structure
data_group = {
"Detectors": [
"Det0",
"Det1",
"Det2",
"Det3",
"Det4",
"Det5",
"Det6",
"Det7",
"Det8",
"Det9",
"Det10",
"Det11",
],
"GroupNb": [1, 2, 2, 3, 3, 4, 1, 1, 2, 4, 1, 2],
"Counts": [2014, 153, 10, 5300, 123, 2000, 1075, 217, 16, 1750, 500, 800],
"RunningOK": [
True,
True,
False,
False,
True,
True,
True,
True,
False,
True,
True,
True,
],
}
df_group = pd.DataFrame(data_group)
# Use 'GroupNb' to group the DataFrame
grouped = df_group.groupby("GroupNb")
for name, group in grouped:
print(f"GroupNb: {name}")
print(group)
GroupNb: 1
Detectors GroupNb Counts RunningOK
0 Det0 1 2014 True
6 Det6 1 1075 True
7 Det7 1 217 True
10 Det10 1 500 True
GroupNb: 2
Detectors GroupNb Counts RunningOK
1 Det1 2 153 True
2 Det2 2 10 False
8 Det8 2 16 False
11 Det11 2 800 True
GroupNb: 3
Detectors GroupNb Counts RunningOK
3 Det3 3 5300 False
4 Det4 3 123 True
GroupNb: 4
Detectors GroupNb Counts RunningOK
5 Det5 4 2000 True
9 Det9 4 1750 True
The DataFrame has been split into 4 groups according to the content of GroupNb.
Below we group the DataFrame according to RunningOK and then we sum the counts of Dets with RunningOK=True.
# Group on value of RunningOK
select_running_det = df_group.groupby(["RunningOK"])
# Sum counts of all running "Det"s
print(
f"Total count of running Dets: {select_running_det.get_group((True, ))['Counts'].sum()}"
)
Total count of running Dets: 8632
# Apply several functions to the groups at once
select_running_det["Counts"].agg(["sum", "mean", "std"])
| sum | mean | std | |
|---|---|---|---|
| RunningOK | |||
| False | 5326 | 1775.333333 | 3052.452347 |
| True | 8632 | 959.111111 | 788.265571 |
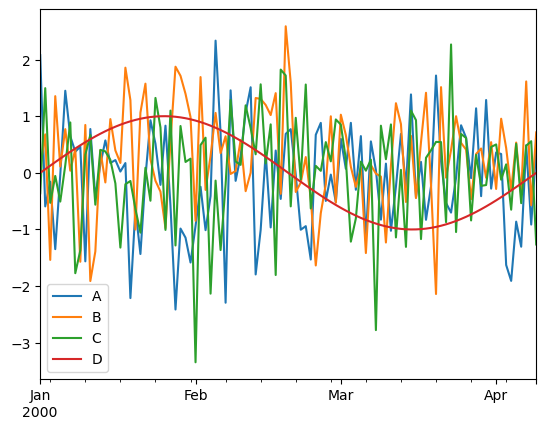
Plotting#
# import matplotlib.pyplot as plt
# %matplotlib widget
df_to_plot = pd.DataFrame(
np.concatenate(
(
np.random.randn(100, 3),
np.sin(np.linspace(0, 2 * np.pi, 100)).reshape(100, 1),
),
axis=1,
),
index=pd.date_range("1/1/2000", periods=100),
columns=["A", "B", "C", "D"],
)
df_to_plot.plot();
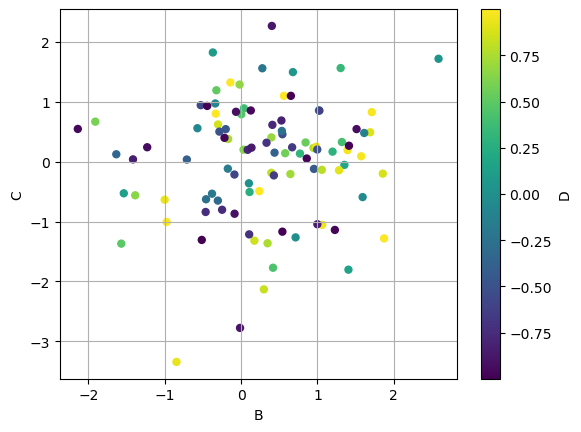
Plotting one column vs. another using a third column to color the points
df_to_plot.plot.scatter(x="B", y="C", c="D", grid=True, s=25);
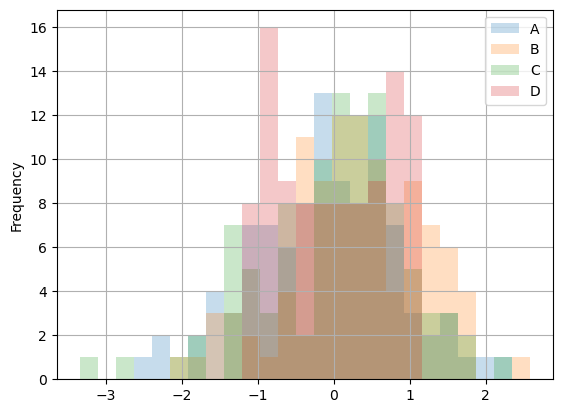
Histograms
Histograms can be plotted using DataFrame.plot.hist() and Series.plot.hist().
help(pd.DataFrame.plot.hist)
Help on function hist in module pandas.plotting._core:
hist(self, by: 'IndexLabel | None' = None, bins: 'int' = 10, **kwargs) -> 'PlotAccessor'
Draw one histogram of the DataFrame's columns.
A histogram is a representation of the distribution of data.
This function groups the values of all given Series in the DataFrame
into bins and draws all bins in one :class:`matplotlib.axes.Axes`.
This is useful when the DataFrame's Series are in a similar scale.
Parameters
----------
by : str or sequence, optional
Column in the DataFrame to group by.
.. versionchanged:: 1.4.0
Previously, `by` is silently ignore and makes no groupings
bins : int, default 10
Number of histogram bins to be used.
**kwargs
Additional keyword arguments are documented in
:meth:`DataFrame.plot`.
Returns
-------
class:`matplotlib.AxesSubplot`
Return a histogram plot.
See Also
--------
DataFrame.hist : Draw histograms per DataFrame's Series.
Series.hist : Draw a histogram with Series' data.
Examples
--------
When we roll a die 6000 times, we expect to get each value around 1000
times. But when we roll two dice and sum the result, the distribution
is going to be quite different. A histogram illustrates those
distributions.
.. plot::
:context: close-figs
>>> df = pd.DataFrame(np.random.randint(1, 7, 6000), columns=['one'])
>>> df['two'] = df['one'] + np.random.randint(1, 7, 6000)
>>> ax = df.plot.hist(bins=12, alpha=0.5)
A grouped histogram can be generated by providing the parameter `by` (which
can be a column name, or a list of column names):
.. plot::
:context: close-figs
>>> age_list = [8, 10, 12, 14, 72, 74, 76, 78, 20, 25, 30, 35, 60, 85]
>>> df = pd.DataFrame({"gender": list("MMMMMMMMFFFFFF"), "age": age_list})
>>> ax = df.plot.hist(column=["age"], by="gender", figsize=(10, 8))
df_to_plot.plot.hist(alpha=0.25, bins=25, grid=True);
DataFrame.hist() plots the histograms of the columns on multiple subplots
df_to_plot.hist(color="k", alpha=0.5, bins=50);
Reading and writing files#
# Create a simple DataFrame to write to files
df_to_write_to_file = pd.DataFrame(
{
"A": ["one", "one", "two", "three"] * 3,
"B": ["A", "B", "C"] * 4,
"C": ["foo", "foo", "foo", "bar", "bar", "bar"] * 2,
"D": np.random.randn(12),
"E": np.random.randn(12),
}
)
df_to_write_to_file
| A | B | C | D | E | |
|---|---|---|---|---|---|
| 0 | one | A | foo | 0.280351 | 0.529483 |
| 1 | one | B | foo | -0.407810 | -1.054804 |
| 2 | two | C | foo | -0.405993 | 0.514961 |
| 3 | three | A | bar | -0.309780 | 0.120868 |
| 4 | one | B | bar | -0.839615 | 0.306638 |
| 5 | one | C | bar | 0.655638 | -1.447115 |
| 6 | two | A | foo | 0.625553 | -0.194368 |
| 7 | three | B | foo | -0.813518 | -0.124487 |
| 8 | one | C | foo | -0.491018 | 0.487561 |
| 9 | one | A | bar | 0.827824 | -0.517703 |
| 10 | two | B | bar | -0.510768 | 0.449158 |
| 11 | three | C | bar | 0.560279 | 0.631953 |
CSV#
# writing a csv file
df_to_write_to_file.to_csv("simple_file.csv")
Options can be added when saving to a .csv file. For example:
Without the index
df_to_write_to_file.to_csv('simple_file.csv', index=False)
Specify a custom delimiter for the CSV output; the default is a comma
df_to_write_to_file.to_csv('simple_file.csv',sep='\t') # Use Tab to separate data
Dealing with missing values
df_to_write_to_file.to_csv('simple_file.csv', na_rep='Unknown') # missing value saved as 'Unknown'
Specifying the precision of the data written to file
df_to_write_to_file.to_csv('simple_file.csv', float_format='%.2f')
Whether to export the column names
df_to_write_to_file.to_csv('simple_file.csv', header=False)
Select columns to be written in the .csv file. Default is None.
df_to_write_to_file.to_csv('simple_file.csv',columns=['C'])
# Reading a csv file
pd.read_csv("simple_file.csv")
| Unnamed: 0 | A | B | C | D | E | |
|---|---|---|---|---|---|---|
| 0 | 0 | one | A | foo | 0.280351 | 0.529483 |
| 1 | 1 | one | B | foo | -0.407810 | -1.054804 |
| 2 | 2 | two | C | foo | -0.405993 | 0.514961 |
| 3 | 3 | three | A | bar | -0.309780 | 0.120868 |
| 4 | 4 | one | B | bar | -0.839615 | 0.306638 |
| 5 | 5 | one | C | bar | 0.655638 | -1.447115 |
| 6 | 6 | two | A | foo | 0.625553 | -0.194368 |
| 7 | 7 | three | B | foo | -0.813518 | -0.124487 |
| 8 | 8 | one | C | foo | -0.491018 | 0.487561 |
| 9 | 9 | one | A | bar | 0.827824 | -0.517703 |
| 10 | 10 | two | B | bar | -0.510768 | 0.449158 |
| 11 | 11 | three | C | bar | 0.560279 | 0.631953 |
HDF5#
An additional library PyTables is required to deal with HDF5 within Pandas. It can be installed within a notebook using
import sys
!{sys.executable} -m pip install tables
# Writing an HDF5 file
# df_to_write_to_file.to_hdf('simple_file.h5', 'df_to_write_to_file', format='table', mode='w')
# Reading an HDF5 file
# pd.read_hdf('simple_file.h5')
Excel#
Dealing with Excel files in Pandas requires openpyxl, xlrd.
To be installed from a notebook:
import sys
!{sys.executable} -m pip install openpyxl xlrd
# Writing an Excel file
# df_to_write_to_file.to_excel('simple_file.xlsx', sheet_name='Sheet1')
# Reading an Excel file
# pd.read_excel('simple_file.xlsx', 'Sheet1')
Exercises#
The solutions can be found in the solutions folder of this repository.
How to combine series to form a dataframe?#
Combine series1 and series2 to form a DataFrame
series1 = pd.Series(["a", "b", "c", "d"])
series2 = pd.Series([1, 2, 3, 4])
Solution:
How to stack two series vertically and horizontally?#
Stack series1 and series2 vertically and horizontally to form a dataframe.
series1 = pd.Series(range(5))
series2 = pd.Series(list("vwxyz"))
Solution:
How to get the positions of items of series A in another series B?#
Get the positions of items of series2 in series1 as a list.
series1 = pd.Series([10, 3, 6, 5, 3, 1, 12, 8, 23])
series2 = pd.Series([1, 3, 5, 23])
Solution:
How to compute difference of differences between consecutive numbers of a series?#
Difference of differences between the consecutive numbers of series.
series = pd.Series([1, 3, 6, 10, 15, 21, 27, 35])
# Desired Output
# Differences: [nan, 2.0, 3.0, 4.0, 5.0, 6.0, 6.0, 8.0]
# Difference of differences: [nan, nan, 1.0, 1.0, 1.0, 1.0, 0.0, 2.0]
Solution:
How to check if a dataframe has any missing values?#
Check if df has any missing values.
df = pd.DataFrame(np.random.randn(6, 4), index=list("abcdef"), columns=list("ABCD"))
df["E"] = [0.5, np.nan, -0.33, np.nan, 3.14, 8]
df
| A | B | C | D | E | |
|---|---|---|---|---|---|
| a | -1.109384 | 0.706269 | 0.224345 | 0.089060 | 0.50 |
| b | -0.332776 | 0.520078 | -0.054311 | -1.703875 | NaN |
| c | 0.427137 | 0.314727 | -2.416297 | -0.089938 | -0.33 |
| d | 0.390647 | 1.807541 | 1.564421 | -1.276061 | NaN |
| e | -0.946299 | -0.732662 | 1.049317 | 0.750035 | 3.14 |
| f | -0.867303 | 0.389175 | -0.552008 | 0.887649 | 8.00 |
Solution:
Playing with groupby and csv files#
load the csv file
biostats.csvto ausersDataFramedetermine the average, minimum and maximum ages per gender
determine the average weight of people over 35 years of age
df = pd.read_csv(
"https://people.sc.fsu.edu/~jburkardt/data/csv/biostats.csv", skipinitialspace=True
)
Solution:
References#
https://pandas.pydata.org/pandas-docs/stable/user_guide/10min.html
Exercises:
https://www.w3resource.com/python-exercises/pandas/index.php
guipsamora/pandas_exercises
CSV files:
https://people.sc.fsu.edu/~jburkardt/data/csv/csv.html