Uniform priors in easyscience#
Bayesian methods are becoming more and more popular in the analysis of model-dependent data, in particular in neutron scattering. Thinking about data probabilistically, helps towards an understanding of Bayesian modelling. Bayesian modelling is about how to include prior knowledge about the system or parameters under study in the analysis.
The same data as previously will be used again.
import numpy as np
np.random.seed(123)
a_true = -0.9594
b_true = 7.294
c_true = 3.102
N = 25
x = np.linspace(0, 10, N)
yerr = 1 + 1 * np.random.rand(N)
y = a_true * x ** 2 + b_true * x + c_true
y += np.abs(y) * 0.1 * np.random.randn(N)
The most straightforward way to include prior knowledge in some analysis is through the use of bounded parameters.
Bounded parameters cannot have values less than some lower bound or greater than some upper bound, as the probability of the parameters having these values is zero.
For example, if the parameter b from the quadratic model has bounds of 0 and 10, then there is an equal probability that the value of b can be anything in between 0 and 10, and a probability of 0 outside those bounds, i.e., it has a uniform prior probability distribution (Fig. 6):
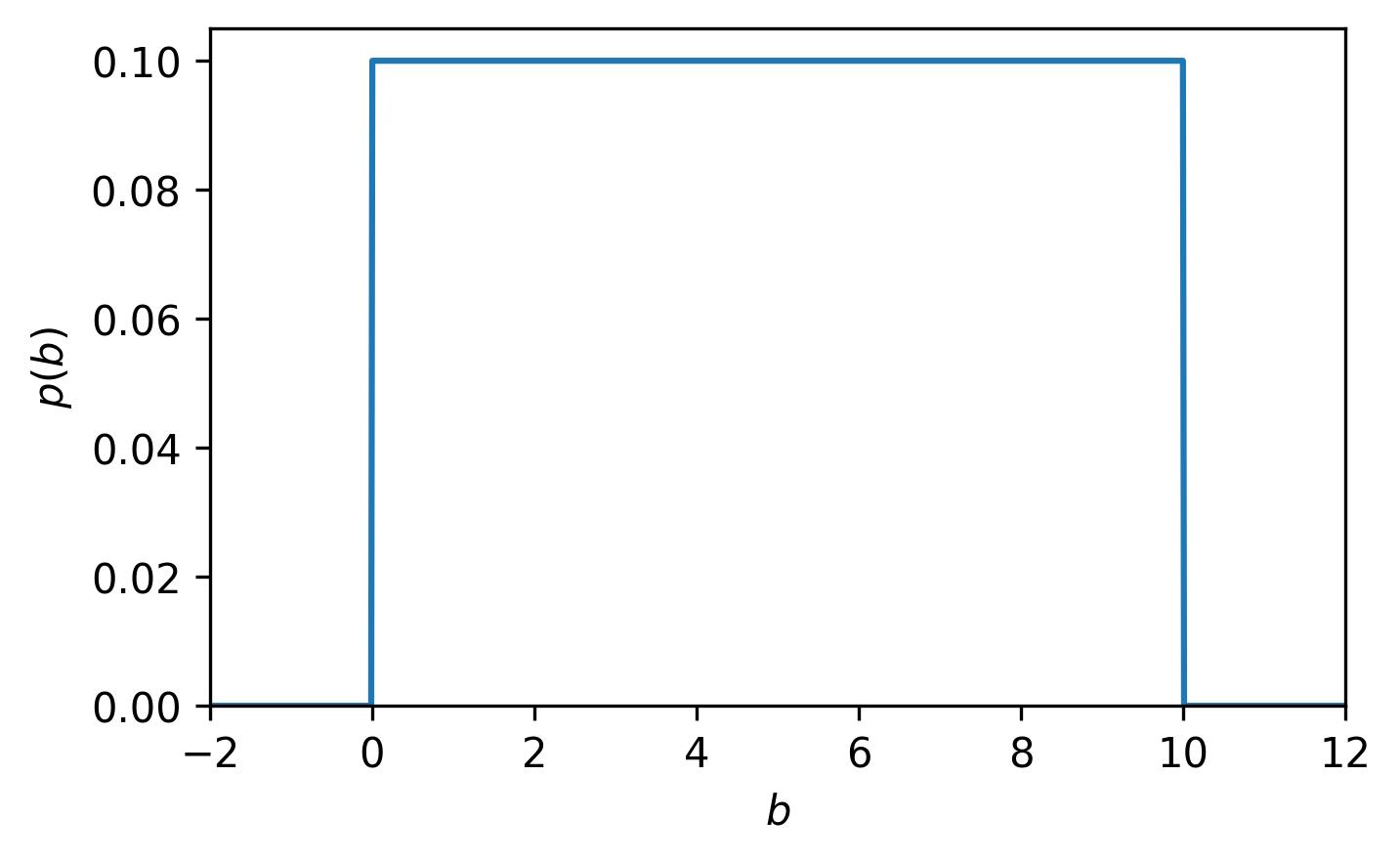
Fig. 6 A normal distribution (blue line), centred on 10.4 with a standard deviation of 1.6.#
Information about bounds can be included in easyscience parameters as min and max values.
from easyscience.Objects.variable import Parameter
a = Parameter(name='a', value=a_true, fixed=False, min=-5.0, max=0.5)
b = Parameter(name='b', value=b_true, fixed=False, min=0, max=10)
c = Parameter(name='c', value=c_true, fixed=False, min=-20, max=50)
We can then perform the analysis in the same fashion as previously, however, this time the bounds will be respected.
from easyscience.Objects.ObjectClasses import BaseObj
from easyscience.fitting import Fitter
def math_model(x, *args, **kwargs):
return a.value * x ** 2 + b.value * x + c.value
quad = BaseObj(name='quad', a=a, b=b, c=c)
f = Fitter(quad, math_model)
res = f.fit(x=x, y=y, weights=yerr)
a, b, c
(<Parameter 'a': -0.9206 ± 0.0373, bounds=[-5.0:0.5]>,
<Parameter 'b': 7.0861 ± 0.3947, bounds=[0.0:10.0]>,
<Parameter 'c': 2.0548 ± 0.8492, bounds=[-20.0:50.0]>)
Currently easyscience only supports uniform prior probability distributions, however, there are plans to extend this in the future.